Calculates the process capability indices cp, cpk, cpkL and cpkU for a given dataset and distribution. A histogram with a density curve is displayed along with the specification limits and a Quantile-Quantile Plot for the specified distribution.
Usage
qcs.pcr(
object,
distribution = c("normal", "beta", "chi-squared", "exponential", "f", "geometric",
"lognormal", "log-normal", "logistic", "t", "negative binomial", "poisson",
"weibull", "gamma"),
limits = c(lsl = -3, usl = 3),
target = NULL,
std.dev = NULL,
boxcox = FALSE,
lambda = c(-5, 5),
confidence = 0.9973,
plot = TRUE,
main = NULL,
...
)Arguments
- object
qcs object of type
"qcs.xbar"or"qcs.one".- distribution
Character string that represent the probability distribution of the data, such as: "normal", "beta", "chi-squared", "exponential", "f", "geometric", "lognormal", "log-normal", "logistic","t", "negative binomial", "poisson", "weibull", "gamma".
- limits
A vector specifying the lower and upper specification limits.
- target
A value specifying the target of the process. If it is
NULL, the target is set at the middle value between specification limits.- std.dev
A value specifying the within-group standard deviation.
- boxcox
Logical value (by default
FALSE). IfTRUE, perform a Box-Cox transformation.- lambda
A vector specifying or numeric value indicating lambda for the transformation.
- confidence
A numeric value between 0 and 1 specifying the nivel for computing the specification limits.
- plot
Logical value indicating whether graph should be plotted.
- main
Title of the plot.
- ...
Arguments to be passed to or from methods.
References
Montgomery, D.C. (1991) Introduction to Statistical Quality Control, 2nd
ed, New York, John Wiley & Sons.
Examples
library(qcr)
data(pistonrings)
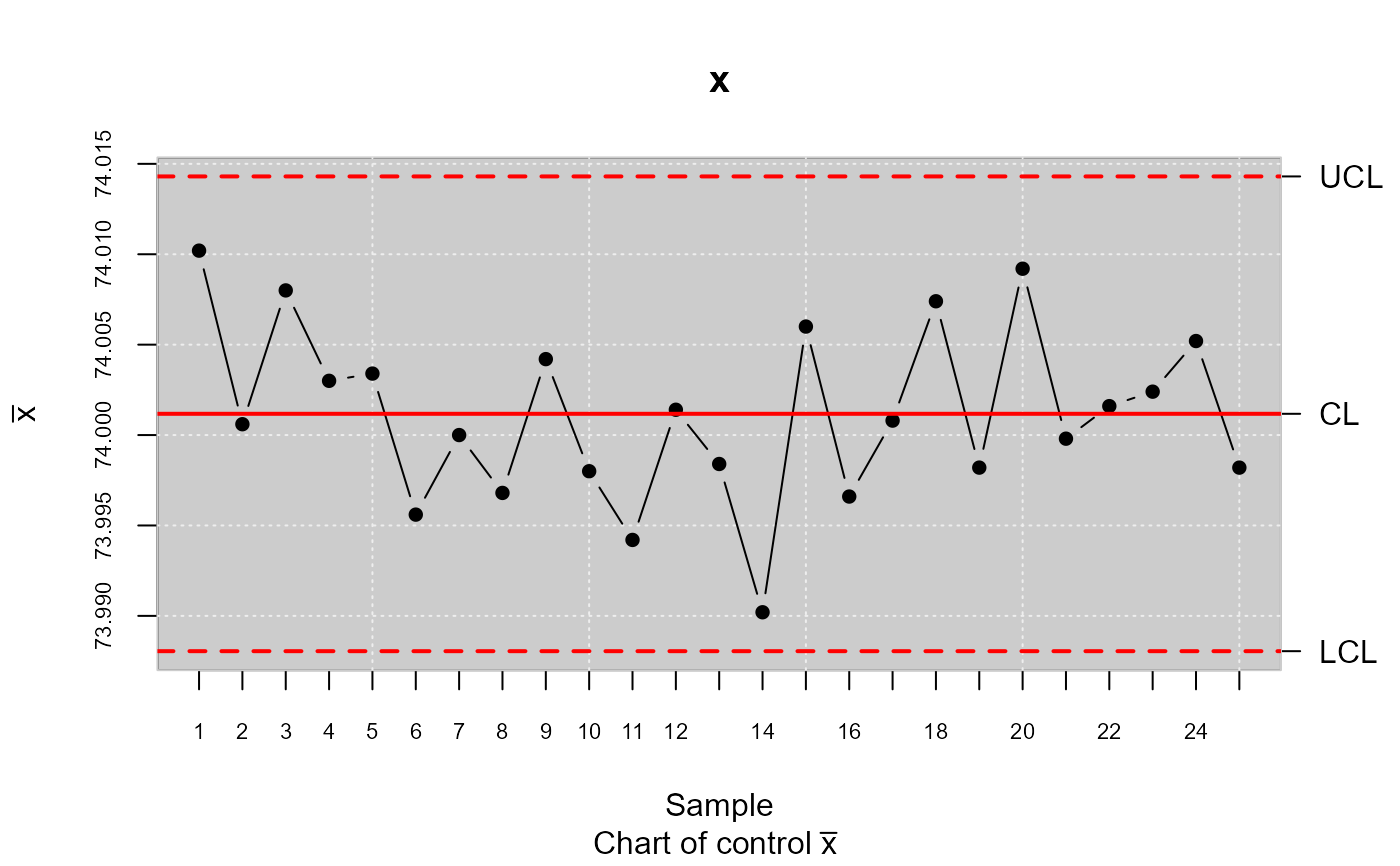
xbar <- qcs.xbar(pistonrings[1:125,],plot = TRUE)
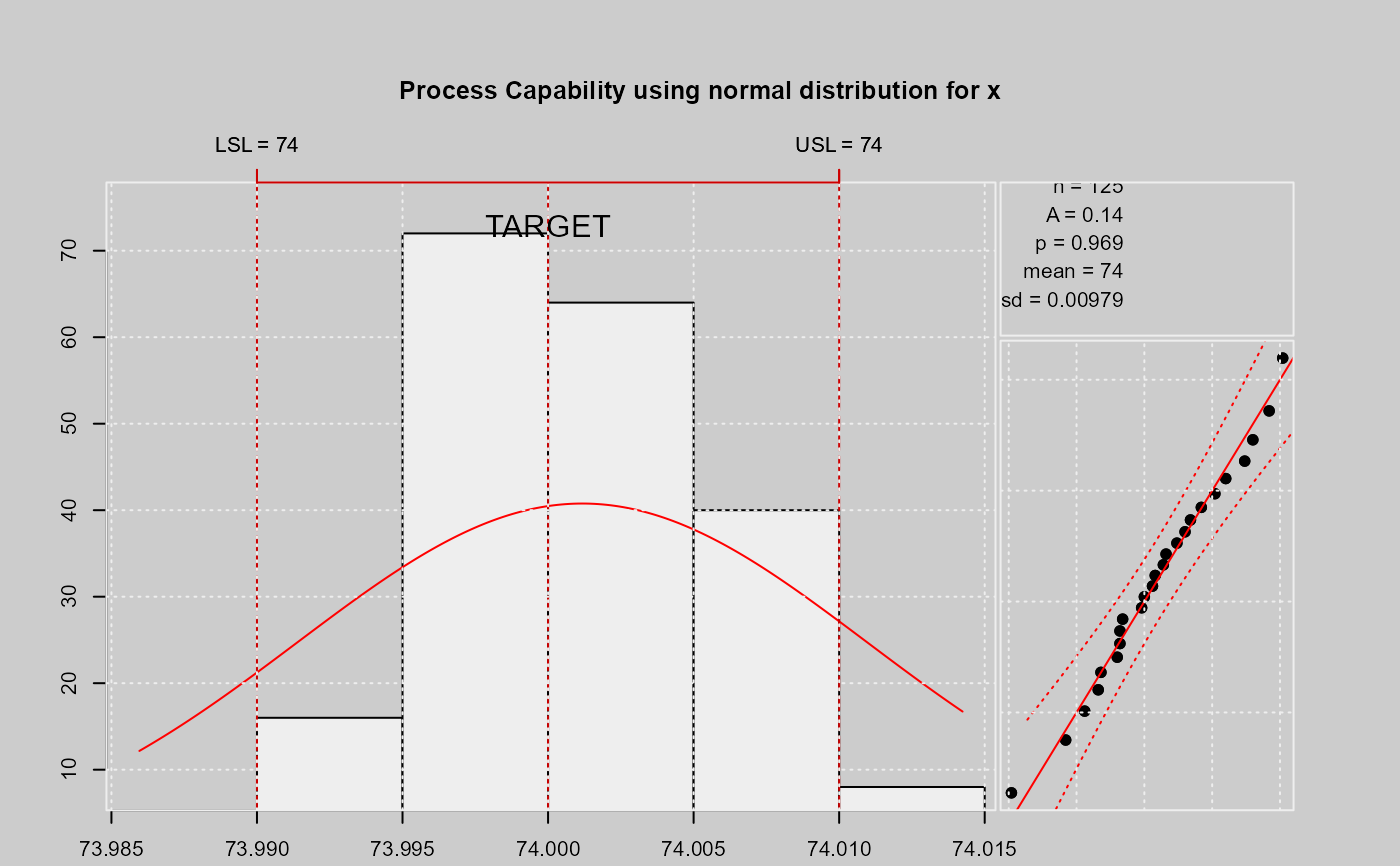
 limits = c(lsl = 73.99, usl = 74.01)
qcs.pcr(xbar, "normal", limits = limits)
#>
#> Process Capability Analysis
#>
#> Call:
#> qcs.pcr(object = xbar, distribution = "normal", limits = limits)
#>
#> Number of obs = 125 Target = 74
#> Center = 74 LSL = 73.99
#> StdDev = 0.009785 USL = 74.01
#>
#> Capability indices:
#>
#> Value
#> Cp 0.3407
#> Cp_l 0.3807
#> Cp_u 0.3006
#> Cp_k 0.3006
#>
#>
#> PPM:
#>
#> Exp<LSL 1.267e+05 Obs<LSL 0
#> Exp>USL 1.836e+05 Obs>USL 1e+12
#> Exp Total 3.103e+05 Obs Total 1e+12
#>
#> Test:
#>
#>
#> Anderson Darling Test for normal distribution
#>
#> data: x
#> Error in round(x$statistic, 4): non-numeric argument to mathematical function
limits = c(lsl = 73.99, usl = 74.01)
qcs.pcr(xbar, "normal", limits = limits)
#>
#> Process Capability Analysis
#>
#> Call:
#> qcs.pcr(object = xbar, distribution = "normal", limits = limits)
#>
#> Number of obs = 125 Target = 74
#> Center = 74 LSL = 73.99
#> StdDev = 0.009785 USL = 74.01
#>
#> Capability indices:
#>
#> Value
#> Cp 0.3407
#> Cp_l 0.3807
#> Cp_u 0.3006
#> Cp_k 0.3006
#>
#>
#> PPM:
#>
#> Exp<LSL 1.267e+05 Obs<LSL 0
#> Exp>USL 1.836e+05 Obs>USL 1e+12
#> Exp Total 3.103e+05 Obs Total 1e+12
#>
#> Test:
#>
#>
#> Anderson Darling Test for normal distribution
#>
#> data: x
#> Error in round(x$statistic, 4): non-numeric argument to mathematical function
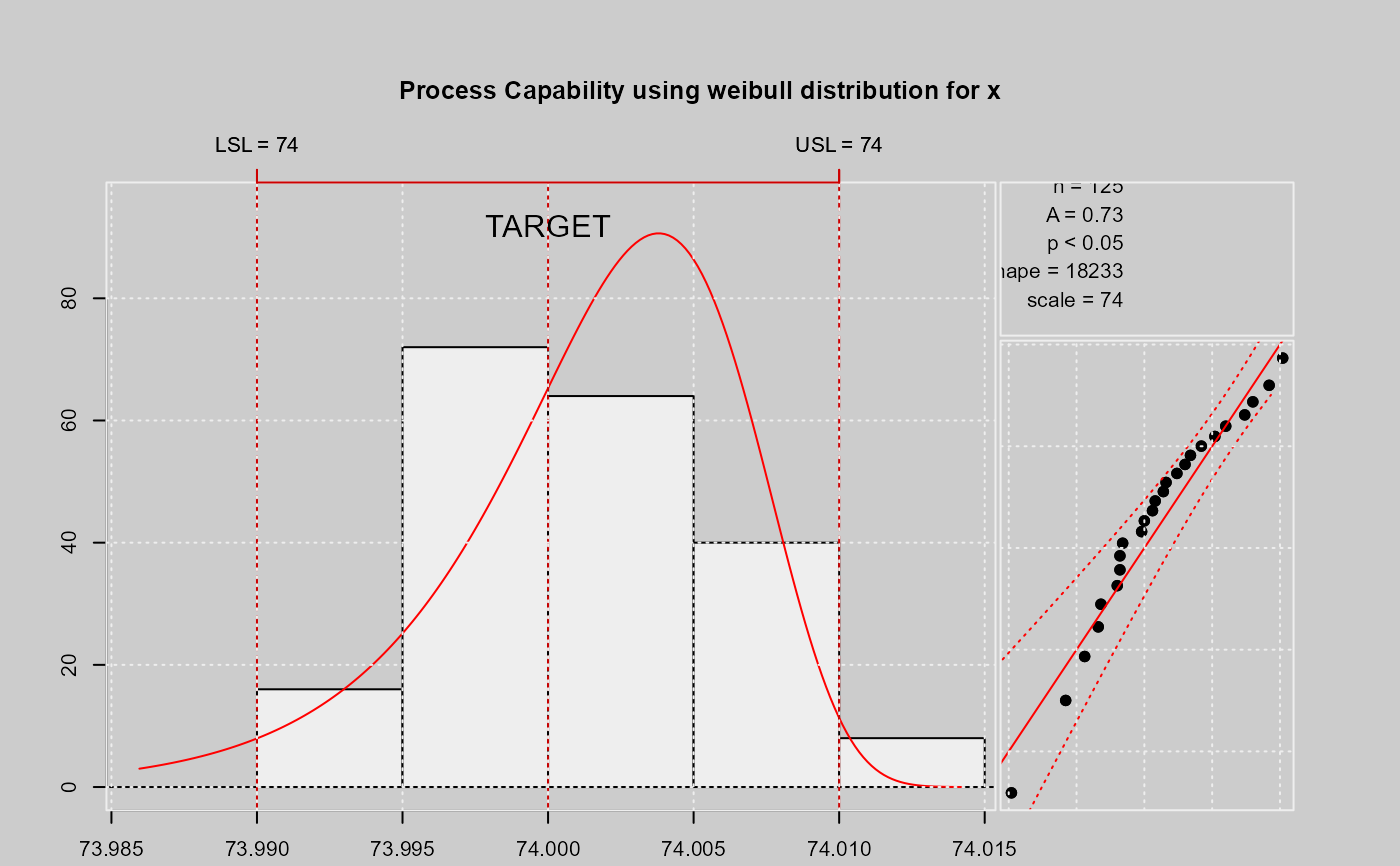
 qcs.pcr(xbar, "weibull", limits = limits)
#>
#> Process Capability Analysis
#>
#> Call:
#> qcs.pcr(object = xbar, distribution = "weibull", limits = limits)
#>
#> Number of obs = 125 Target = 74
#> Center = 74 LSL = 73.99
#> StdDev = 0.009785 USL = 74.01
#>
#> Capability indices:
#>
#> Value
#> Cp 0.5801
#> Cp_l 0.4861
#> Cp_u 0.8402
#> Cp_k 0.4861
#>
#>
#> PPM:
#>
#> Exp<LSL 3.283e+04 Obs<LSL 0
#> Exp>USL 9965 Obs>USL 1e+12
#> Exp Total 4.279e+04 Obs Total 1e+12
#>
#> Test:
#>
#>
#> Anderson Darling Test for weibull distribution
#>
#> data: x
#> Error in round(x$statistic, 4): non-numeric argument to mathematical function
qcs.pcr(xbar, "weibull", limits = limits)
#>
#> Process Capability Analysis
#>
#> Call:
#> qcs.pcr(object = xbar, distribution = "weibull", limits = limits)
#>
#> Number of obs = 125 Target = 74
#> Center = 74 LSL = 73.99
#> StdDev = 0.009785 USL = 74.01
#>
#> Capability indices:
#>
#> Value
#> Cp 0.5801
#> Cp_l 0.4861
#> Cp_u 0.8402
#> Cp_k 0.4861
#>
#>
#> PPM:
#>
#> Exp<LSL 3.283e+04 Obs<LSL 0
#> Exp>USL 9965 Obs>USL 1e+12
#> Exp Total 4.279e+04 Obs Total 1e+12
#>
#> Test:
#>
#>
#> Anderson Darling Test for weibull distribution
#>
#> data: x
#> Error in round(x$statistic, 4): non-numeric argument to mathematical function